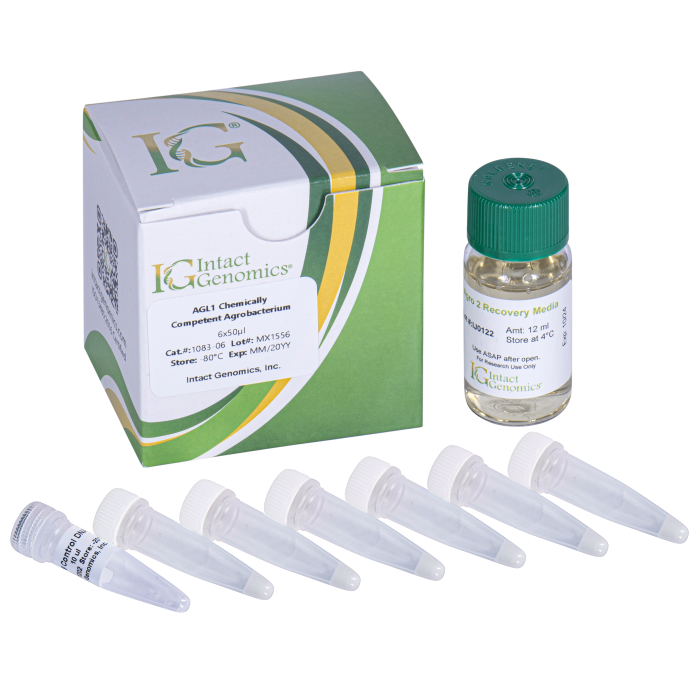
AGL1 Chemically Competent Cells allow for Agrobacterium-mediated transformation of Arabidopsis thaliana as well as maize and other monocots.
AGL1 Agrobacterium Chemically Competent Cells
Price range: $204.00 through $541.00
Description
Intact Genomics Agrobacterium tumefaciens AGL1 (AGL-1) cells are optimized for the highest transformation efficiencies which are ideal for applications requiring high transformation efficiencies, such as with cDNA or gDNA library construction.
The AGL1 strain has a C58 chromosomal background that carries an insertion mutation in its recA recombination gene which stabilizes recombinant plasmids. It also carries rifampicin and carbenicillin resistance in its genome for selection. AGL1 contains the Ti plasmid pTiBO542 from which the T-DNA region sequences have been deleted. Transformation with a binary vector containing the missing T-region results in a functional T-DNA binary system that allows for the transfer of genetic material into a host plant’s genome. Therefore, this system is often used for the Agrobacterium-mediated transformation of Arabidopsis thaliana as well as maize and other monocots.
Specifications
Competent cell type: Chemically Competent
Species: A. tumefaciens
Strain: AGL1
Format: Tubes
Transformation efficiency: ≥ 1 x 105 cfu/µg pCAMBIA1391z DNA
Blue/white screening: No
Shipping condition: Dry ice
Reagents Included
- AGL1 Agrobacterium Chemically Competent Cells
- DNA (pCAMBIA1391z, 500 pg/µl)
- Recovery medium
Note: Liquid nitrogen is required.
Storage
AGL1 Agrobacterium Chemically Competent Cells: -80 ºC
pCAMBIA1391z control DNA: -20 ºC
Recovery medium: 4 ºC
Quality Control
Transformation efficiency is tested by using the pCAMBIA1391z control DNA supplied with the kit and using the protocol in this manual. Transformation efficiency should be ≥1 x 105 CFU/µg pCAMBIA1391z DNA. Untransformed cells are tested for appropriate antibiotic sensitivity.
General Guidelines
Follow these guidelines when using AGL1 Agrobacterium Chemically Competent Cells:
- Handle competent cells gently as they are highly sensitive to changes in temperature or mechanical lysis caused by pipetting.
- Thaw competent cells on ice, and transform cells immediately following thawing. After adding DNA, mix by tapping the tube gently. Do not mix cells by pipetting or vortexing.
Calculation of Transformation Efficiency
Transformation Efficiency (TE) is defined as the number of colony-forming units (cfu) produced by transforming 1µg of the plasmid into a given volume of competent cells.
TE = Colonies/µg/Plated
Transform 1 µl of (500 pg/µl) pCAMBIA1391z control plasmid into 50 µl of cells, and add 950 µl of Recovery Medium. Recover for 3 hours and plate 100 µl. Count the colonies on the plate in two days. If you count 5 colonies, the TE is calculated as follows:
Colonies = 5
µg of DNA = 0.0005
Dilution = 100/1000 = 0.1
TE = 5/.0005/.1 = 1×105
Please note, all agrobacterial strains are not well-studied for antibiotic resistance and there are many agrobacterial strains. Therefore, it is the customer’s responsibility to make sure his/her vectors are compatible with the Agrobacterial strains if he/she uses an alternate antibiotic selection than kanamycin-selection.1082-06 1082-18 1083-10
Additional information
| µl | 6×50µl, 10×50µl, 18×50µl |
|---|
Transformation Protocol
Use this procedure to transform AGL1 Chemically Competent Agrobacterium cells . Do not use these cells for electrocompetent transformation.
1) Place microcentrifuge tubes on ice.
2) Remove competent cells from the -80 °C freezer and thaw completely on wet ice (10-15 minutes).
3) Aliquot 1 µl ( 10pg -1 µg) of DNA to the chilled microcentrifuge tubes on ice.
4) When the cells are thawed, add 50μl of cells to each DNA tube on ice and mix gently by tapping 4-5 times. For the pCAMBIA1391z control, add 1 µl of (500 pg/µl) DNA to the 50 µl of cells on ice. Mix well by tapping. Do not pipette up and down or vortex to mix, this can harm cells and decrease transformation efficiency.
5) Keep tubes on ice for 5 minutes, and then transfer to liquid nitrogen for 5 minutes.
6) Incubate tubes for additional 5 minutes in 37°C water bath.
7) Immediately add 950µl of Recovery Medium or any other medium of choice to the tube, pipette up and down three times to re-suspend the cells.
8) Incubate tubes at 30 °C for 3 hours at 200 RPM.
9) Dilute the cells as appropriate then spread 20-200 μl cells onto a pre-warmed selective plate. For the pCAMBIA1391z control, you may plate 100 μl of undiluted transformation mix onto a YT plate containing 15 μg/ml rifampicin and 50 μg/ml kanamycin. Use a sterilized spreader or autoclaved ColiRoller™ plating beads to spread evenly.
10) Incubate the plates for 2 – 3 days at 30 °C.