Fungal Artificial Chromosome (FAC)
Fungal Artificial Chromosome (FAC) –
Novel drug, anti-fungi, and insecticide discovery pipeline from fungi*
Fungi are prolific producers of secondary metabolites (SMs) and have been producing 45% of bioactive molecules from all microbial sources since the turn of the century. Conservative estimates suggest that there are more than 5 million fungal species (Blackwell, 2011), of which fewer than 5% have been described and less than 1% are available in the world’s culture collections (Colwell, 1997). In addition, each of these fungal genomes may harbor 50 or more different SM gene clusters (Norberg et al., 2013). Fungal secondary metabolite (SM) gene cluster sequences available for characterization far outstrip our current ability to characterize each cluster, as most of the fungi are not genetically amendable. To address this post-genomic SM characterization gridlock, we have demonstrated a new technology that generates unbiased shuttle bacterial artificial chromosome (BAC) or fungal artificial chromosome (FAC) libraries, pooled FAC next-gen sequences to rapidly capture all the SM gene clusters in a fungal genome, exhibited heterologous expression in suitable host systems, and characterized high-throughput chemical analysis pipelines.
Construction of unbiased Random Shear shuttle BAC or FAC libraries of fungi with average insert 100 kb or larger
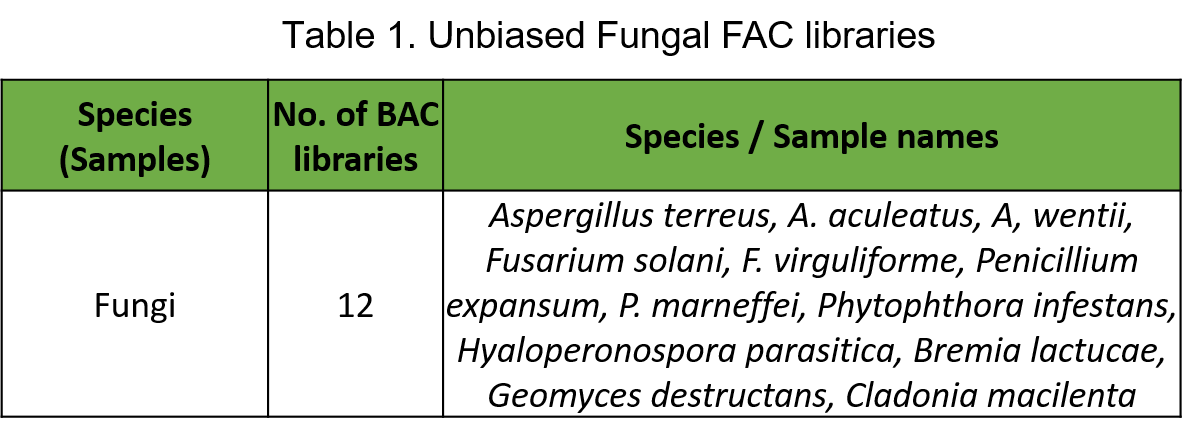
Scientists at Intact Genomics have successfully constructed unbiased Random Shear shuttle bacterial artificial chromosome (BAC or FAC) libraries of 12 fungal species or strains with average insert sizes of at least 100 kb (Table 1).
Identify the entire set of intact SM gene clusters containing FACs from fungi (A FAC = intact SM gene cluster)
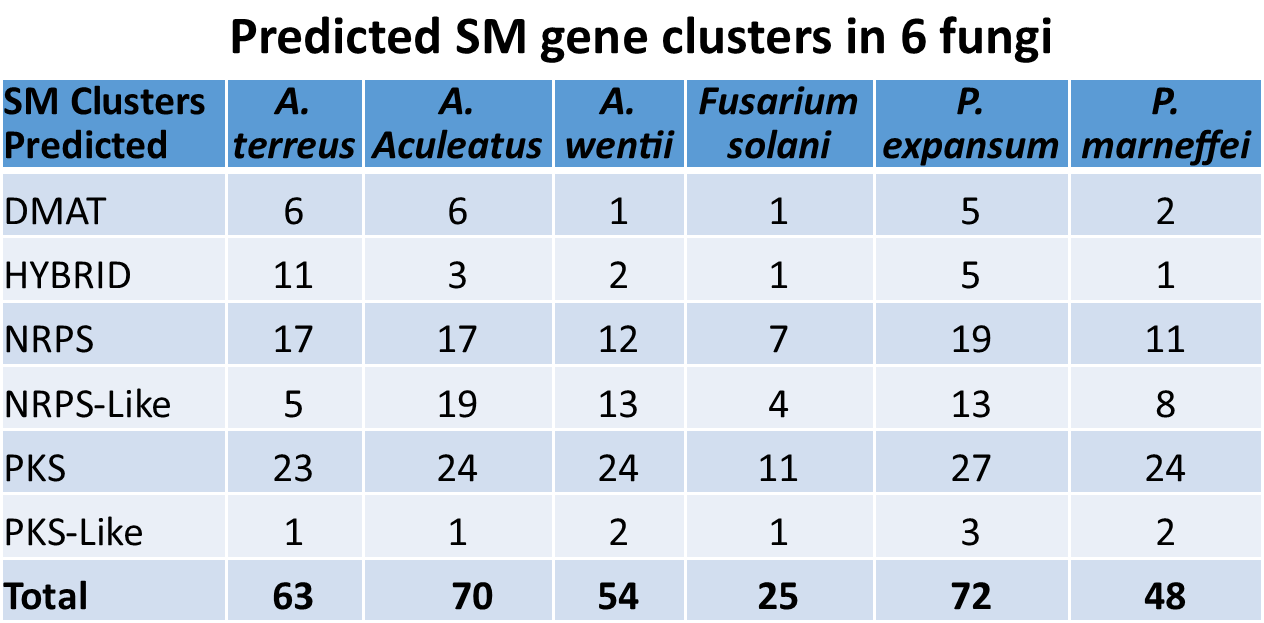
We have successfully constructed unbiased FAC libraries of 6 sequenced fungal species with average insert sizes of at least 100 kb for natural product discovery. The number of large secondary metabolite (SM) gene clusters from each of the 6 fungal genomes was predicted ranging from 25-72 with the size ranging from ~20 kb up to more than 100 kb (Table 2). By either PCR-based FAC library screening or FAC end sequencing- genome sequence alignment, intact SM gene clusters are readily to be identified: Figure 1a, b show the examples of astechrome gene cluster (6J7) and putative gene cluster 23 (9D19) respectively. We have identified FAC clones containing more than 200 intact SM gene clusters from the 6 fungi, one FAC equals an intact SM gene cluster.
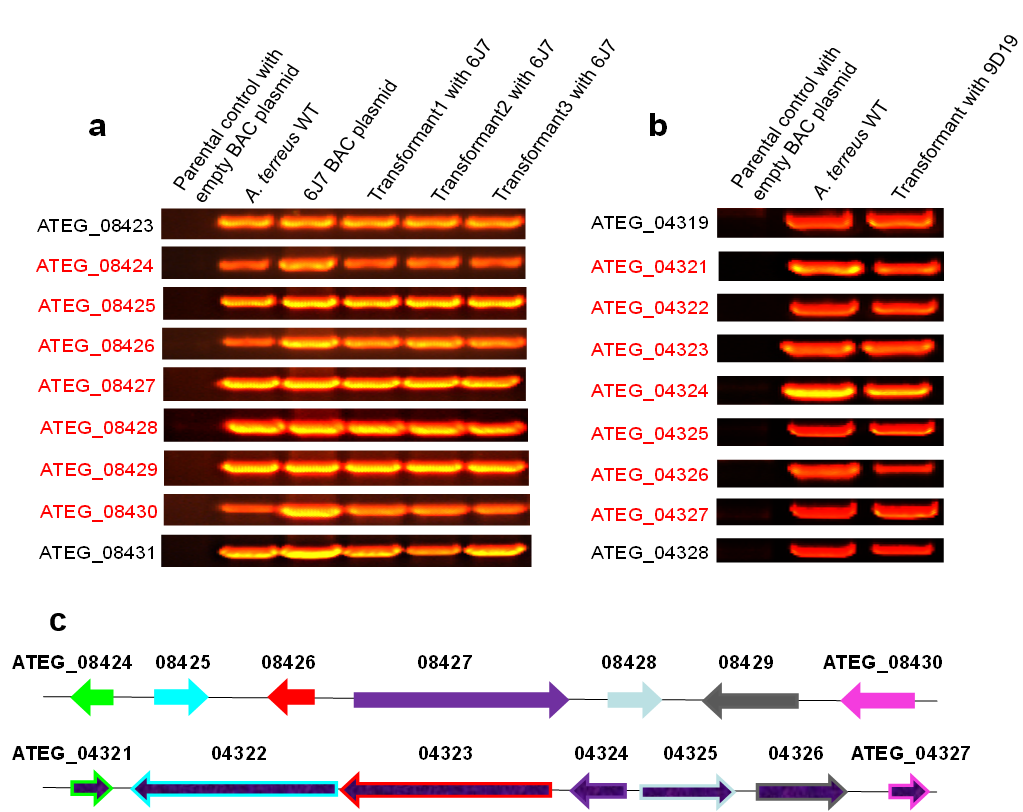
Figure 1. FAC and FAC transformants contain intact gene clusters confirmed by PCR. Panel a: Astechrome gene cluster (6J7) is present in A. nidulans transformed with FAC as examined by PCR. The seven astechrome genes (red) are not present in wild type A. nidulans but are in A. terreus and all A. nidulans 6J7 transformants. Panel b: Putative gene cluster 23 (9D19) is present in A. nidulans transformed with FAC 9D19 as examined by PCR. The seven cluster genes (red) are not present in wild type A. nidulans but are in A. terreus and A. nidulans 9D19 transformant. Panel c shows both Astechrome gene cluster (6J7) on the top and the cluster 23 (9D19) on the bottom.
Figure 2
Functional study with unbiased BACs in fungi
An overview of fungal shuttle BAC or FAC technology is presented in Figure 2. More than 200 intact SM gene clusters up to 200 kb from 6 sequenced fungal species were individually captured in a self-replicating fungal shuttle BAC system. Candidate BACs were successfully shuttled between E. coli and the heterologous expression host A. nidulans. For example, the BAC A. nidulans transformant or FAC strain was characterized in a novel liquid chromatography-high resolution mass spectrometry (LC-HRMS) and data analysis pipeline leading to the discovery of the A. terreus astechrome biosynthetic machinery (Figure 3). We have also developed the techniques to precisely engineer and activate cryptic SM clusters and at least one each of novel compound-derivatives of antibacterial, anticancer, antifungal, and anti-insecticide has been discovered in our pipeline (data not shown). The novel technology enables drug, antifungi, and insecticide discovery in large scale not possible before in microbes, metagenomes, and especially in filamentous fungi
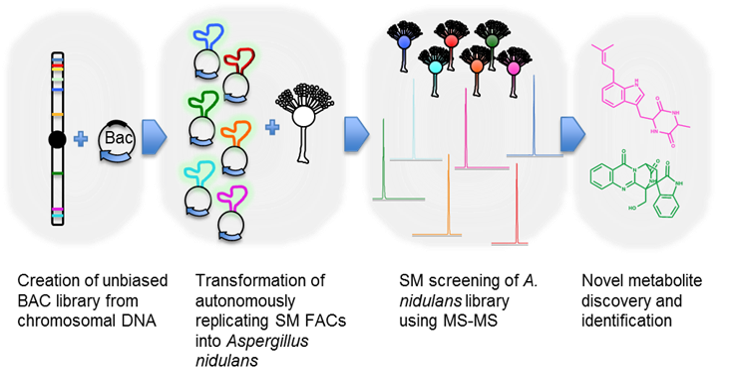
Figure 3. A diagram showing the workflow of cryptic SM production
High molecular weight (HMW) genomic DNA from sequenced fungi was mechanically sheared and cloned into shuttle AMAI-BAC (or FAC) clones, SM FACs were selected and transformed into A. nidulans, transformants were screened for metabolites, followed by compound identification and structure elucidation.
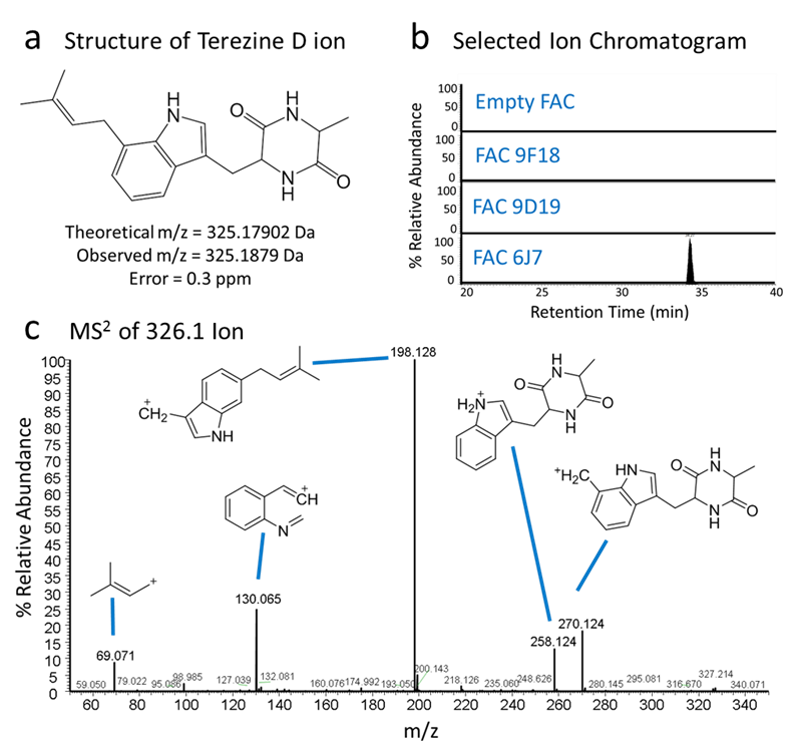
Figure 4. Identification of natural products and structure elucidation of compounds. (a) A compound was identified in FAC 6J7 cell extracts with exact mass matching the theoretical exact mass of terezine D, the predicted product of FAC 6J7. (b) Selected ion chromatograms of lysates from cells containing FAC 6J7 and all other analyzed FACs show that the compound of interest is unique to the FAC 6J7 expressing strain. (c) Analysis of MS2 fragmentation data for the identified compound lead to identification of predicted fragments of terezine D, confirming the compound’s identity.
Scientists at Intact Genomics and our collaborators have developed a breakthrough technology (fungal artificial chromosome, FAC, a patent pending), which has potential to revive natural product discovery and new thinking of synthetic biology for agriculture, pharmaceutical, bioenergy, environmental protection, and many other industries. The innovative FAC technology is capable to capture 50,000~1,000,000 full-length fungal SM pathways, enable precisely silent SM pathway editing, and access ONE million fungal SM compounds. Similarly Intact Genomics has developed a shuttle BAC technology enabling the capture of 50,000~1,000,000 full-length SM pathways from bacteria and soil metagenomes (microbiome).
* The technologies developed were partially supported by the NIH SBIR programs, and the FAC technology has been developed by the collaboration with Professor Nancy P. Keller’s lab at Department of Medical Microbiology and Immunology and Bacteriology, University of Wisconsin – Madison, and Professor Neil L. Kelleher’s lab at Department of Chemistry, Northwestern University.
