DNA Library Screening
Colony picking, re-arraying, and duplicating
Intact Genomics offers custom colony picking, re-arraying and duplicating, either as a stand-alone service or as part of our library construction services (Random Shear or conventional BAC or fosmid libraries).
Intact Genomic’ BAC/fosmid libraries are delivered as clones frozen in 96- or 384-well plates. Colonies are first grown on an agar tray and then robotically picked into plates for further growth and storage. We use colony picking robots to automatically transfer colonies from a solid to liquid medium in the 96- or 384-well plates. The robot selects colonies according to roundness, size and color via a build-in camera before picking. Our well-trained engineers and technicians achieve the highest robot picking accuracy and guarantee a library array without empty wells.
High-Accuracy
Robotically selecting the right colonies, automatic colony picking
Fast Delivery
Can pick up to 30,000 selected colonies per day from trays or petri dishes
High-Quality
High-quality control system, zero empty wells guaranteed

ig® 3D DNA Pools (SuperPools and Plate-Row-Column Pools) Intact Genomics’ BAC Library SuperPools and Pools system provides a platform for simple and efficient identification of specific clones. With this system, researchers can screen an entire BAC library by two consecutive rounds of PCR to localize the clone(s) of interest to specific well(s).
A standard Intact Genomics BAC DNA SuperPool contains 10 library plates. However, the number of plates for each SuperPool can be varied to meet the customer’s needs (for example, 8-15 plates/SuperPool).
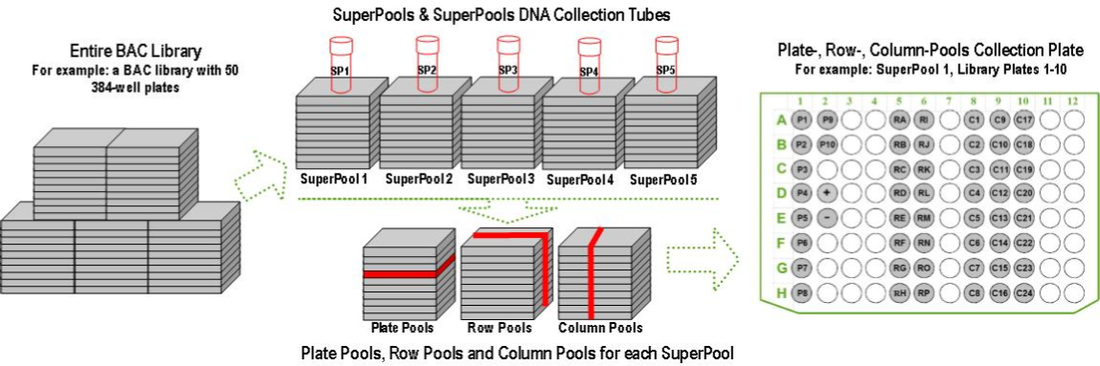
The BAC library is divided into SuperPools. Aliquots of the samples from each SuperPool are combined in various ways to form Plate-Pools, Row-Pools, and Column-Pools, which are provided on a 96-well Plate-Row-Column (P-R-C) Pools DNA Collection plate. Each SuperPool contains all the clones’ DNA from 10 consecutively numbered 384-well plates from the BAC library. Each SuperPool has corresponding Plate-Pools, Row-Pools, and Column-Pools, all of which are provided on a P-R-C Pools DNA Collection Plate (Figure 3). Screening the P-R-C Pools plate by PCR will identify the specific plate, row, and column of the well that contains the sequence of interest.
3D DNA pools for library screening
Find your targeted clone in as few at 51 PCR reactions
No radioactive materials needed
Fast, simple and accurate
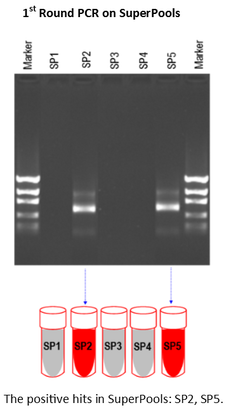
1st Round PCR Screening of SuperPools
The 1st Round of PCR is used to determine which of the SuperPools contains positive wells. Â In this example, positive signals were seen in SuperPools SP2 and SP5 (Figure 4).
2nd Round PCR Screening of SuperPools
The 2nd Round PCR screening of SuperPool SP2 involves analysis of the 50 reactions plus positive and negative controls of the P-R-C Pools plate from this SuperPool: 10 Plate-Pools plus positive and negative control PCRs, 16 Row-Pools, and 24 Column-Pools.
To facilitate screening large numbers of DNA clones, Intact Genomics can robotically print viable, bacterial cells from 384-well plates onto high-density, 22.2×22.2 cm or 8×12 cm nylon filters. After overnight growth of the clones, the filters are processed. As a result, DNA from each clone is permanently fixed to a known position on the filter, allowing rapid screening using standard DNA hybridization procedures. Positive clones can then be identified and retrieved from the original plates. Each nylon filter can be screened 3-5 times. Intact Genomics provides 22.2×22.2 cm or 8×cm filters printed at a choice of two densities. All clones are printed in duplicate. We can also provide customized filters to meet your needs, for example, different spotting patterns, single or double spotting, etc.
Filters prepared for you to hybridize
Automation ensures accuracy and quality of the arrays
We offer high-quality BAC/Fosmid end sequencing at a low price. Up to 700 bp per run, minimum of 90% successful reads. From DNA preparation to data analysis, you only need to provide BAC/fosmid clones for us.