Description
Exonuclease III (Exo III) is an exodeoxyribonuclease that digests one strand of duplex DNA from a blunt end, 5′ overhang or nick (1, 2). The enzyme acts in a 3′→5′ direction producing stretches of single-stranded DNA (1, 2). Exo III is not active on 3′ overhangs of 4 or more bases in length and ssDNA (2). The enzyme has also RNase H, 3′-DNA phosphatase, and apurinic DNA endonuclease activities (2, 3). Exo III digestions proceed at a uniform rate yielding predictable and reproducible results. However, the observed rate of nucleotide excision can vary depending on the individual characteristics of the reaction mix such as reaction temperature, ionic strength, template base content, template helical structure, and enzyme-to-DNA ratios (3-5).
Applications
• 3′ → 5′ exonuclease
• Production of unidirectional nested deletions
• Production of intermediates for site-directed mutagenesis
• Preparation of strand-specific probes
• Preparation of single-stranded substrates for dideoxy sequencing
Protein purity
The physical purity of this enzyme is ≥99% as assessed by SDS-PAGE with Coomassie® blue staining (see figure below).
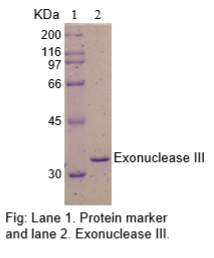
Product Source
E. coli BL21 (DE3) strain expressing E. coli Exonuclease III gene.
Product Includes
- Exonuclease III
- 10x Exonuclease III reaction buffer
1x Exonuclease III reaction buffer
33 mM Tris-acetate, 66 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT, pH 7.5 @ 25ºC
Storage Buffer
50 mM Tris-HCl, 50 mM KCl, 1 mM DTT, 0.1 mM EDTA, 50% Glycerol, pH 7.5 @ 25ºC
Storage Temperature
–20ºC
Heal inactivation
70°C for 20 min
Unit Definition
One unit is defined as the amount of enzyme that incorporates 1 nmoles of dNTP into acid-insoluble form in 30 minutes at 37º C with duplex DNA.
Quality Control assays
Exonuclease III is free from contaminating nuclease activities.
References
- Weiss, B. (1976) J. Biol. Chem. 251, 1896.
- Rogers, S.G. and Weiss, B. (1980) Meth. Enzymology 65, 201.
- Sambrook, J. et al., (1989) in: Molecular Cloning: A Laboratory Manual (2nd ed.), Cold Spring Harbor Laboratory Press, New York.
- Richardson, C.C. et al., (1964) J. Biol. Chem. 239, 251.
- Guo, L.H. and Wu, R. (1982) Nucl. Acids Res. 10, 2065
3412 3415