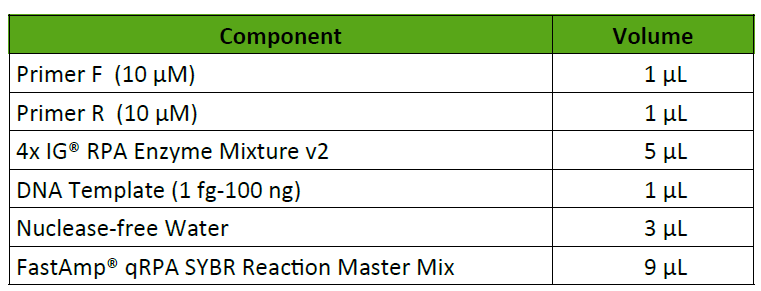
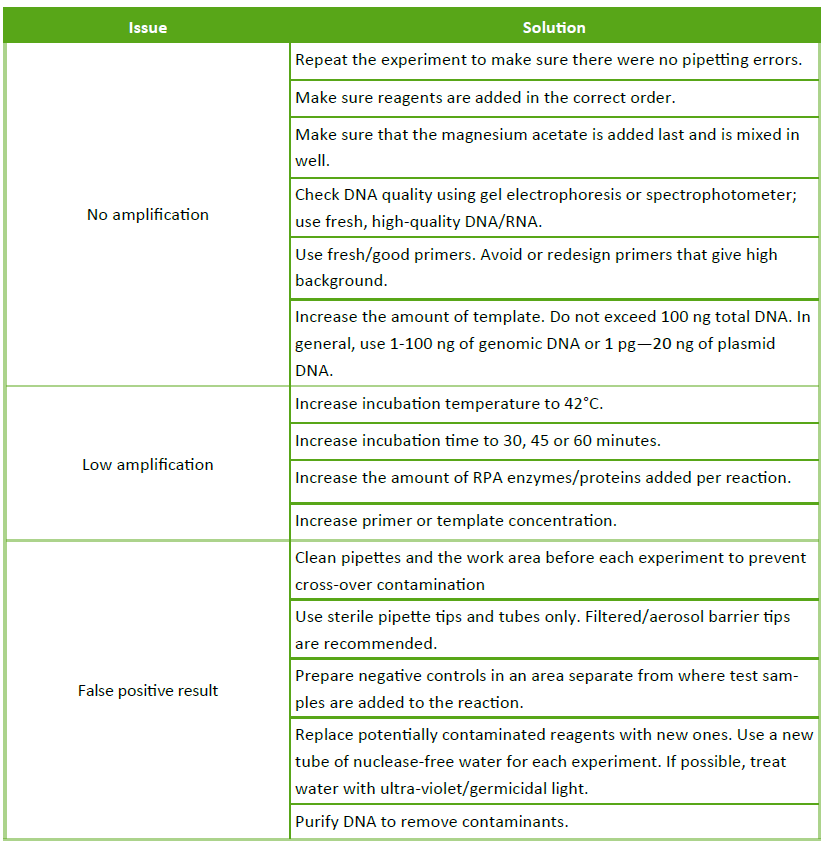
Description
IG® scientists engineered a novel FastAmp® qRPA SYBR kit that contains master mixes with ready-to-use cocktails containing all components except primers and template for the amplification, detection, and quantification of DNA in RPA. With our new technology, RPA progress can be monitored in real-time and quantified using standardized template concentrations. The FastAmp® qRPA SYBR kit is compatible with high throughput (96-well or 384-well) qPCR thermocyclers without temperature cycles for faster results, i.e. the experiment is complete in just 30 minutes at constant 37⁰C.
A key feature of the FastAmp® qRPA SYBR Kit is that it can be used to screen primer conditions for the lowest possible Non-template control (NTC) background. Most polymerases can produce erroneous byproducts with short DNA strands that result in the appearance of a non-specific NTC signal1. These byproducts are also capable of binding the intercalating SYBR dye, but products occur with lower probability than specific primer-target annealing and take longer to amplify. Over time the formation of byproducts eventually can lead to a background signal in both PCR and RPA. Using this knowledge to our advantage, we show that it is possible to screen for well-behaved primers that have ultra-low SYBR background relative to the specific, intended signal generated (Figure 1).
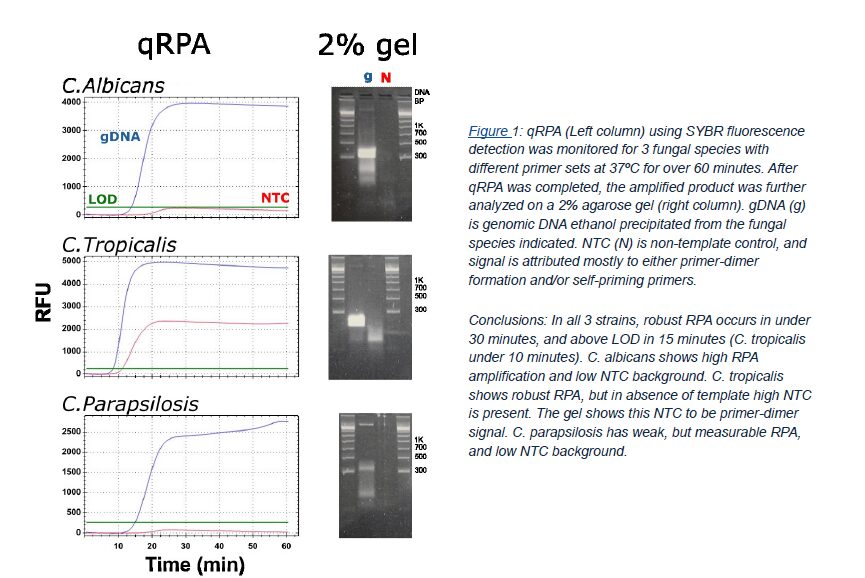
RPA/qRPA Primer Design:
While RPA generally favors longer primers—usually more than 20 base pairs—we find that virtually any primer design software has a similar success rate between RPA and PCR if a simple tool is used to avoid strong primer dimerization and strong primer secondary structure. For RPA primer design, we suggest designing 8 to 10 primers per forward and reverse direction. For most targets, it is unnecessary to test every possible primer combination. For example, by screening all reverse primers against a single forward primer, picking the best reverse primer and then using it to screen all the forward primers, a good primer-pair can be found in 16 to 20 reactions.
Kit Includes:
- FastAmp® qRPA SYBR Reaction Master Mix
- IG® RPA Enzyme Mixture v2 (4x)
- 280 mM Magnesium Acetate (MgOAc)
- Nuclease-free Water
** The mix also includes SYBR® Green I fluorescent dye, ROX dye, MgCl2, dNTPs and stabilizers. The use of ROX dye is necessary for all Applied Biosystems instruments and is optional for the Stratagene Mx3000P Mx3005P and Mx4000 cyclers. Bio-Rad, Qiagen,
Eppendorf, Illumina and Roche instruments do not require ROX dye. The concentration of ROX Dye is (5 nM) for this product.
Storage Temperature: -20°C
Quality Control:
- All component proteins of the Enzyme Mixture are free from detectable nuclease activities.
- RPA activity of each lot is validated to be consistent with prior IG® lots and to be equal to or better than major competitors in the market.
References:
- Cromie GA, Connelly JC, Leach DR (2001) Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell 8: 1163–1174
- Michel B, Grompone G, Flores MJ, Bidnenko V (2004) Multiple pathways process stalled replication forks. Proc Natl Acad Sci U S A 101: 12783–12788
- Liu J, Ehmsen KT, Heyer WD, Morrical SW (2011) Presynaptic filament dynamics in homologous recombination and DNA repair. Crit Rev Biochem Mol Biol 46: 240–270
Related Products:
- T4 UvsX Recombinase (Cat.# 3562) , Glycerol-free T4 UvsX DNA Recombinase (Cat.# 3562GF)
- T4 gp32 Protein (Cat.# 3515), Glycerol-free T4 gp32 Protein (Cat.# 3513GF)
- T4 UvsY Protein (Cat.# 3572), Glycerol-free T4 UvsY Protein (Cat.# 3572GF)
- Bsu DNA Polymerase (Cat.# 3585), Glycerol-free Bsu (Cat.# 3585GF)
- ig® Recombinase Polymerase Amplification (RPA) Kit v2 (Cat.# 3533, 3534, 3536)
- Exonuclease IV (Nfo) (Cat.# 3425)
3611 3615 3617


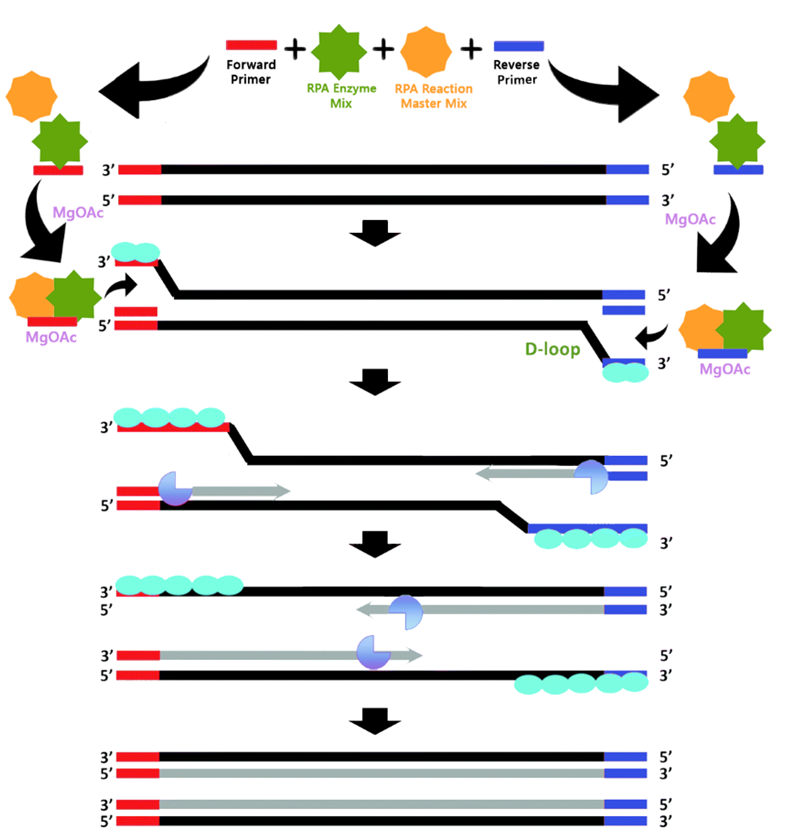 Recombinase Polymerase Amplification (RPA) Technology:
Recombinase Polymerase Amplification (RPA) Technology: